dimspy.process package¶
Submodules¶
dimspy.process.mass_calibration module¶
dimspy.process.peak_alignment module¶
Cluster and align peaklists into peak matrix.
New in version 0.1.
-
dimspy.process.peak_alignment.align_peaks(peaks, ppm=2.0, block_size=2000, fixed_block=True, edge_extend=10, ncpus=None)¶ Cluster and align peaklists into a peak matrix.
Parameters: - peaks – list of peaklists for alignment
- ppm – the hierarchical clustering cutting height, i.e., ppm range for each aligned mz value. Default = 2.0
- block_size – number peaks in each centre clustering block. This can be a exact or approximate number depends on the fixed_block parameter. Default = 2000
- fixed_block – whether the blocks contain fixed number of peaks. Default = True
- edge_extend – ppm range for the edge blocks. Default = 10
- ncpus – number of CPUs for parallel clustering. Default = None, indicating using as many as possible
Return type: PeakMatrix object
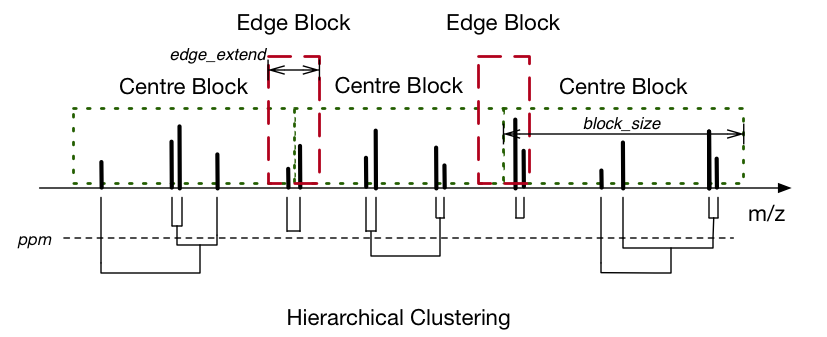
This function uses hierarchical clustering to align the mz values of the input peaklists. The alignment “width” is decided by the parameter of ppm. Due to a large number of peaks, this function splits them into blocks with fixed or approximate length, and clusters in a parallel manner on multiple CPUs. When running, the edge blocks are clustered first to prevent separating the same peak into two adjacent centre blocks. The size of the edge blocks is decided by edge_extend. The clustering of centre blocks is conducted afterwards.
After merging the clustering results, all the attributes (mz, intensity, snr, etc.) are aligned into matrix accordingly. If multiple peaks from the same sample are clustered into one mz value, their attributes are averaged (for real value attributes e.g. mz and intensity) or concatenated (string, unicode, or bool attributes). The flag attributes are ignored. The number of these overlapping peaks is recorded in a new intra_count attribute matrix.
dimspy.process.peak_filters module¶
PeakList and PeakMatrix filters.
New in version 0.1.
-
dimspy.process.peak_filters.filter_attr(pl, attr_name, max_threshold=None, min_threshold=None, flag_name=None, flag_index=None)¶ Peaklist attribute values filter.
Parameters: - pl – the target peaklist
- attr_name – name of the target attribute
- max_threshold – maximum threshold. A peak will be unflagged if the value of it’s attr_name is larger than the threshold. Default = None, indicating no threshold
- min_threshold – Minimum threshold. A peak will be unflagged if the value of it’s attr_name is smaller than the threshold. Default = None, indicating no threshold
- flag_name – name of the new flag attribute. Default = None, indicating using attr_name + ‘_flag’
- flag_index – index of the new flag to be inserted into the peaklist. Default = None
Return type: PeakList object
This filter accepts real value attributes only.
-
dimspy.process.peak_filters.filter_ringing(pl, threshold, bin_size=1.0, flag_name='ringing_flag', flag_index=None)¶ Peaklist ringing filter.
Parameters: - pl – the target peaklist
- threshold – intensity threshold ratio
- bin_size – size of the mz chunk for intensity filtering. Default = 1.0 ppm
- flag_name – name of the new flag attribute. Default = ‘ringing_flag’
- flag_index – index of the new flag to be inserted into the peaklist. Default = None
Return type: PeakList object
This filter will split the mz values into bin_size chunks, and search the highest intensity value for each chunk. All other peaks, if it’s intensity is smaller than threshold x the highest intensity in that chunk, will be unflagged.
-
dimspy.process.peak_filters.filter_mz_ranges(pl, mz_remove_rngs, flag_name='mz_range_remove_flag', flag_index=None)¶ Peaklist mz range filter.
Parameters: - pl – the target peaklist
- mz_remove_rngs – the mz ranges to remove. Must be in the format of [(mz_min1, mz_max2), (mz_min2, mz_max2), …]
- flag_name – name of the new flag attribute. Default = ‘mz_range_remove_flag’
- flag_index – index of the new flag to be inserted into the peaklist. Default = None
Return type: PeakList object
This filter will remove all the peaks whose mz values are within any of the ranges in the mz_remove_rngs.
-
dimspy.process.peak_filters.filter_rsd(pm, rsd_threshold, qc_label='qc', flag_name='rsd_flag')¶ PeakMatrix RSD filter.
Parameters: - pm – the target peak matrix
- rsd_threshold – threshold of the RSD of the QC samples
- qc_label – tag label to unmask qc samples
- flag_name – name of the new flag. Default = ‘rsd_flag’
Return type: PeakMatrix object
This filter will calculate the RSD values of the QC samples. A peak with a QC RSD value larger than the threshold will be unflagged.
-
dimspy.process.peak_filters.filter_fraction(pm, fraction_threshold, within_classes=False, class_tag_type=None, flag_name='fraction_flag')¶ PeakMatrix fraction filter.
Parameters: - pm – the target peak matrix
- fraction_threshold – threshold of the sample fractions
- within_classes – whether to calculate the fraction array within each class. Default = False
- class_tag_type – tag type to unmask samples within the same class. Default = None, indicating untyped tags
- flag_name – name of the new flag. Default = ‘fraction_flag’
Return type: PeakMatrix object
This filter will calculate the fraction array over all samples or within each class (based on class_tag_type). The peaks with a fraction value smaller than the threshold will be unflagged.
-
dimspy.process.peak_filters.filter_blank_peaks(pm, blank_label, fraction_threshold=1, fold_threshold=1, method='mean', rm_blanks=True, flag_name='blank_flag')¶ PeakMatrix blank filter.
Parameters: - pm – the target peak matrix
- blank_label – tag label to mask blank samples
- fraction_threshold – threshold of the sample fractions. Default = 1
- fold_threshold – threshold of the blank sample intensity folds. Default = 1
- method – method to calculate blank sample intensity array. Valid values include ‘mean’, ‘median’, and ‘max’. Default = ‘mean’
- rm_blanks – whether to remove (not mask) blank samples after filtering
- flag_name – name of the new flag. Default = ‘blank_flag’
Return type: PeakMatrix object
This filter will calculate the intensity array of the blanks using the “method”, and compare with the intensities of the other samples. If fraction_threshold% of the intensity values of a peak are smaller than the blank intensities x fold_threshold, this peak will be unflagged.
dimspy.process.scan_processing module¶
-
dimspy.process.scan_processing.remove_edges(pls_sd)¶
-
dimspy.process.scan_processing.read_scans(fn, source, function_noise, min_scans=1, filter_scan_events=None)¶
-
dimspy.process.scan_processing.average_replicate_scans(ID, pls, ppm=2.0, min_fraction=0.8, rsd_thres=30.0, block_size=2000, ncpus=None)¶
-
dimspy.process.scan_processing.join_peaklists(ID, pls)¶