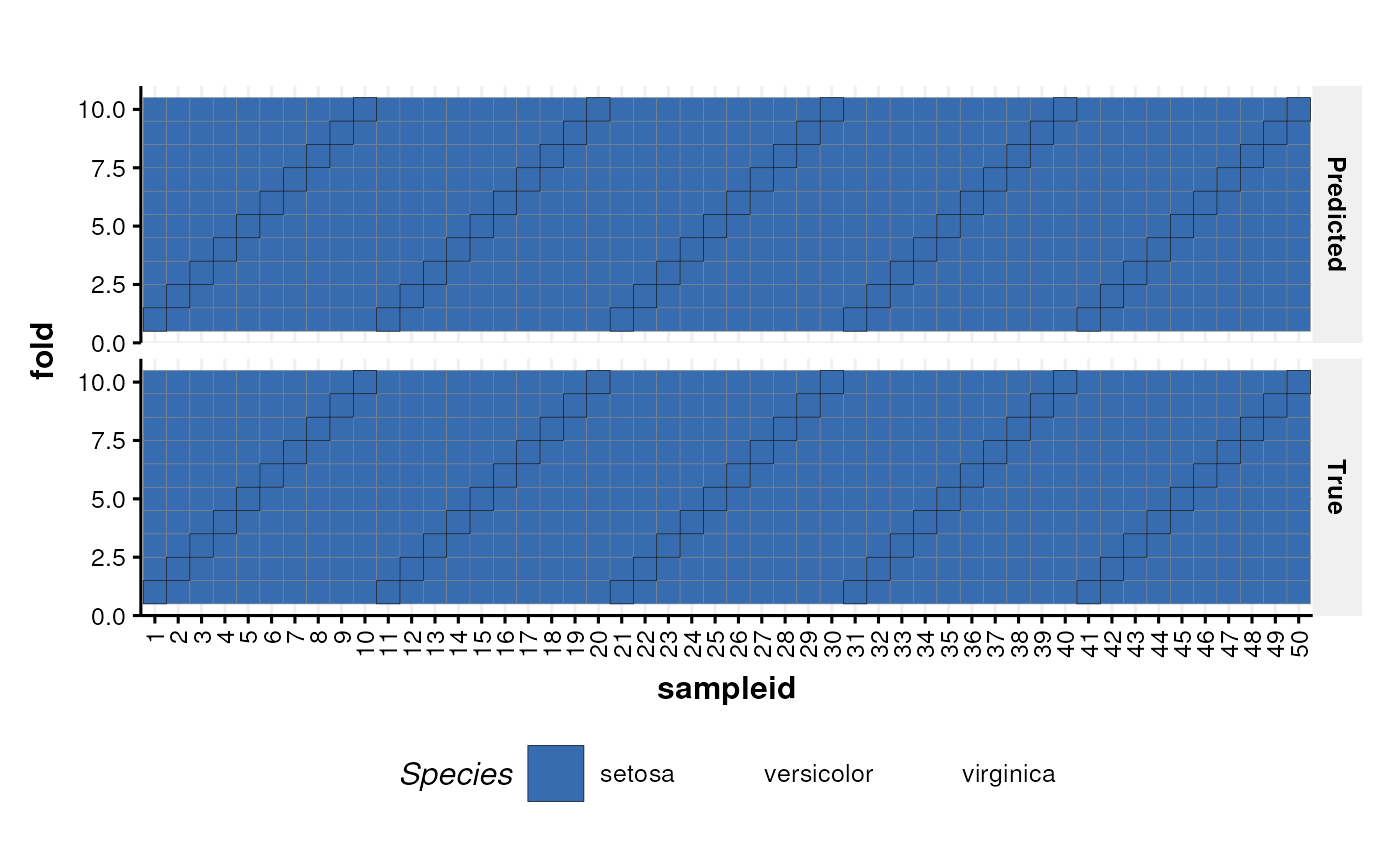
A graphic for visualising the true class and the predicted class of samples in all groups for all cross-validation folds.
Value
A
kfoldxcv_grid
object. This object has no output slots.
See chart_plot in the struct package to plot this chart object.
Inheritance
A kfoldxcv_grid object inherits the following struct classes: [kfoldxcv_grid] >> [chart] >> [struct_class]
Examples
M = kfoldxcv_grid(
factor_name = "V1",
level = "level_1")
D = iris_DatasetExperiment()
I = kfold_xval(factor_name='Species') *
(mean_centre() + PLSDA(factor_name='Species'))
I = run(I,D,balanced_accuracy())
C = kfoldxcv_grid(factor_name='Species',level='setosa')
chart_plot(C,I)
#> Warning: Removed 900 rows containing missing values or values outside the scale range
#> (`geom_tile()`).